Tongue cancer is the most predominant form of oral cancer in developed countries with varying incidence in developing countries wherein etiological factors such as chewing betel-quid comprising betel leaf, areca nut and slaked lime is a part of the tradition. While several large-scale genome-sequencing efforts of advanced stage primary oral tumors have been described, systematic efforts to catalogue somatic alterations in tobacco/ nut chewing associated early stage (pT1/2) HPV-negative tongue tumors has been lacking.
In a comprehensive effort to characterize tongue tumors derived from Indian patients:
- First, we performed functional analysis of biological relevant alterations of 3 primary tumor-derived oral cancer cell lines (AW13516, AW8507 and NT8E) established from Indian head and neck cancer patients (by other groups at the Centre) using integrated genomic approach. The findings establish mutant nuclear receptor binding protein NRBP1 as a novel oncogene in oral cancer [2].
- Second, using in house developed HPVDetector [1], we established that unlike Caucasian population, tongue tumors of Indian origin display significantly low HPV infectivity (n=46), suggesting an alternative mode underlying the disease;
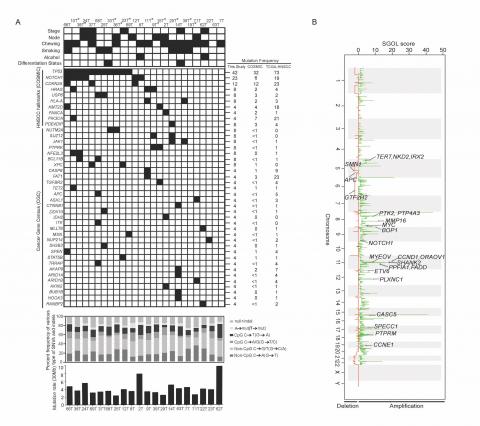
- Thirdly, we characterized the somatic alterations underlying the tongue cancer genome of 57 tobacco/nut chewing HPV-negative early stage tongue tumors by whole exome (n=47) and transcriptome (n=17) sequencing [5]. Post depleting variants against in-house developed TMC-SNPdb, the study revealed a classical tobacco mutational signature C:G>A:T transversion in 53% patients [3]. Of significance, unlike TCGA HNSCC data set, NOTCH1 harbors significantly lower frequency of inactivating mutations (4%), is somatically amplified and overexpressed in 31% and 37% of early stage TSCC patients, respectively;
- Fourth, to functionally validate the relevance of Notch pathway, oral-cancer cell lines with higher basal NOTCH1 expression are enriched in stem cell markers and form spheroids. Inhibition of NOTCH activation by gamma secretase inhibitor or shRNA mediated knockdown of NOTCH1 could inhibit spheroid forming capacity, transformation, survival and migration of the HNSCC cells suggesting an oncogenic role of NOTCH1 in TSCC, thus suggesting NOTCH1 as a therapeutic target [4]; and,
- Finally, an integrated gene-expression analysis of 253 tongue tumor identify upregulation of metastases related pathways and over-expression of MMP10 in 48% tumors along with concomitant downregulation of a miR-944 that is associated with poor prognosis [5].
Figure: Identification of somatic mutations and DNA copy number changes in HPV-negative early stage TSCC tumors.
(a) Mutational features of 25 early tongue squamous carcinoma samples (b) Somatic DNA copy number changes identified using Exome sequencing data. Somatic DNA copy number gains and losses were generated using Segments-of-Gain-Or-Loss (SGOL) scores across 23 TSCC patients. Representative amplified and deleted regions are annotated for HNSCC-associated genes and denoted by an arrow. (Oral Oncology. 2017).