Infectious Pathogen Detector (IPD) can be executed as:
Using the command line:
$ cd /path/to/InfectiousPathogenDetector/
There are 2 modes in the IPD command-line interface based on the sequencing type selected.
For short read data:
$ python3 IPD_cli.py long -h
For long read data:
$ python3 IPD_cli.py short -h
Using the GUI interface
1. Open the terminal and move to the base folder of IPD.
2. Enter:
$ python3 IPD_gui.py
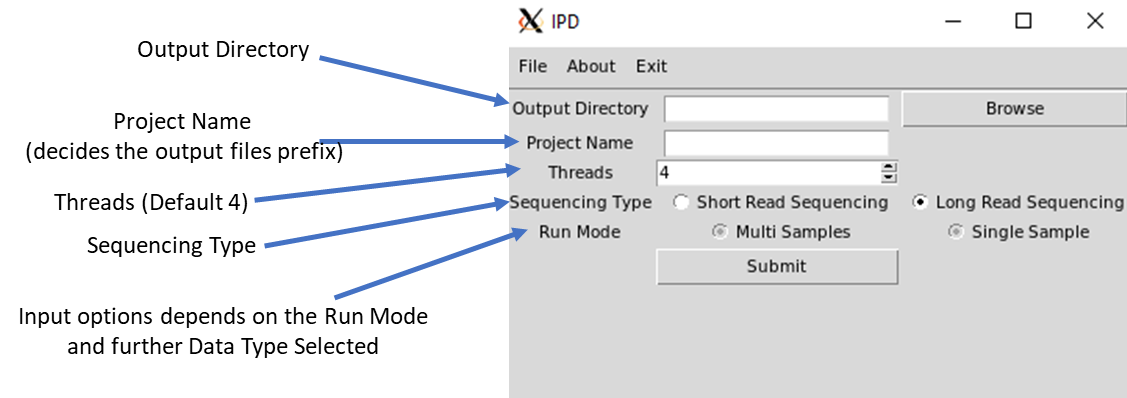
IPD graphical user interface is developed for the analysis of both long and short read to detect the abundance of pathogens and the variants present in them. Following are the entries that are required to be filled before proceeding the run:
1. Output Directory to store the results.
2. Project Name.
3. Number of threads to use.
4. Run Mode.
5. IPD can be run in a multi-sample or a single sample mode.
i. For Multi-sample run mode, user need
to provide a sample info file containing tab separated fastq files (in case of Paired-end
sample, tab separated R1 and R2 fastq file with path) and sample name. Sample name and
Project name will be used as prefix for all the output files.
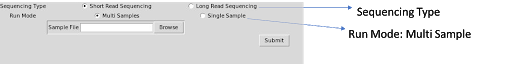
ii. For Single sample run mode there is further two options for the data type, paired-end and
single-end. It enables the user to browse the fastq input files. Project name will be used as the
prefix for all the output files in this case.

Infectious Pathogen Detector Report:
IPD Report has the following sections:
1. Basic Alignment Statistical summary: It includes total reads, aligned reads and read length of each sample in the project.
2. Per Base Coverage for SARS-CoV2 : The read depth of each base of SARS-CoV2 genome is calculated and log2 of the reads is taken and sample-wise plots are generated
3. Relative Abundance: Stack-bar plot illustrates the relative abundance of Human, Pathogen, SARS-CoV2 and unaligned reads for each sample. The FPKM values of SARS-CoV2 are plotted in the adjacent bar plot.
4. Novel SARS-CoV2 Variants: Annotated variants not present in the IPD SARS-CoV2 vcf-database used are tabulated.
5. Variant Based SARS-CoV2 Clade Assignment: Based on the mutational profile of the sample’s clade assessment is done and tabulated in the last section of the report.
Apart from the HTML SARS-CoV2 report, IPD generates other tabulated output which are as follows:
1. Finalcount.tsv: it contains all the raw and normalised counts of all 1060 pathogen included in the database.
2. Final_anno.vcf: It contains the annotated variants for all the pathogens present in database.