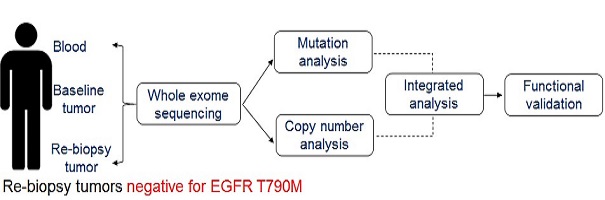
Discovery of therapeutically relevant alterations in human genome – a functional genomics approach
Researcher:Dr. Pratik Chandrani
Funding:
DBT-Wellcome, Terry Fox foundation, Tata Memorial Centre
Collaborators:
Dr. Kumar Prabhash, Tata Memorial Hospital
Lung cancer remains the leading cause of cancer related deaths across the globe. According to the Globocan, around 2 million new lung cancer cases were reported and the numbers of deaths due to lung cancer were more than 1.7 million contributing to 18.6% of all cancer related deaths in 2018. In India, lung cancer constitutes 6.9 per cent of all new cancer cases and 9.3 per cent of all cancer related deaths in both sexes, it is the commonest cancer and cause of cancer related mortality in men. Our pioneering work (involving a large cohort of 1000 odd samples) establishes that while EGFR mutations are present in over 30% of East Asian and 10% of Caucasian lung adenocarcinoma patients, they are only found in about 23-25% of Indian lung adenocarcinoma patients (PLoS One 2013 Apr; PLoS One 2013 Oct; Ind J Cancer 2013; F1000Res 2015). Similarly, KRAS mutations are present at 60% lower frequency in Indian lung adenocarcinoma patients than compared to the Caucasian population (Br J Cancer 2014). Our more recent work establishes landscape of actionable mutations beyond EGFR and KRAS in Indian lung cancer patients (Annals of Oncology 2016). Taken together, these study forms a crucial basis to rationalize targeted therapy in India and to adopt genetic testing in a clinical diagnostic laboratory at an affordable pricing—as the genetic test developed in collaboration with Dr. Kumar Prabhash and Dr. Anuradha Choughule to genotype EGFR mutations has been able to remarkably reduce its cost from ~$200 to $12 per test (!) that is now being offered at the Tata Memorial Hospital on routine basis—enabling and transforming the way EGFR mutation profiling is been carried out at an affordable cost for every lung cancer patient. Our other seminal contribution from ACTREC includes an in-depth functional characterization of FGFR1 and FGFR3 alterations as novel candidate therapeutic targets in lung cancer (PLoS One 2011; Annals of Oncology 2016). Using elegant genetic, biochemical and mouse-xenograft based mechanistic characterization led to the discovery of FGFR3 activating mutations in lung adenocarcinoma patients of Indian origin. Our work thus opens a possibility for cancer treatment.
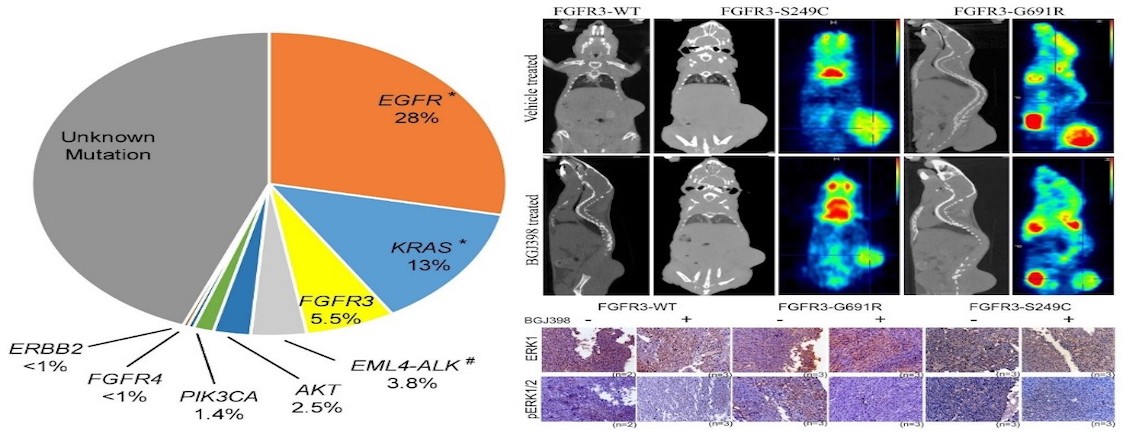
Figure: Drug-sensitive FGFR3 mutations in lung adenocarcinoma. Left panel reflect first comprehensive landscape of therapeutically relevant alterations in lung aenocarcinomapatients of Indian origin. Right upper panel shows that the novel FGFR3 mutations derived from lung cancer patients forms FGFR inhibitor sensitive tumors; lower panel shows the drug is specific. (Annals of Oncology. 2017).