Running HPVDetector in GUI mode:
HPV Detector can be executed with GUI by running HPVdetector_GUI from Linux/UNIX shell:
> ./HPVdetector_GUI
Or
> bash HPVdetector_GUI
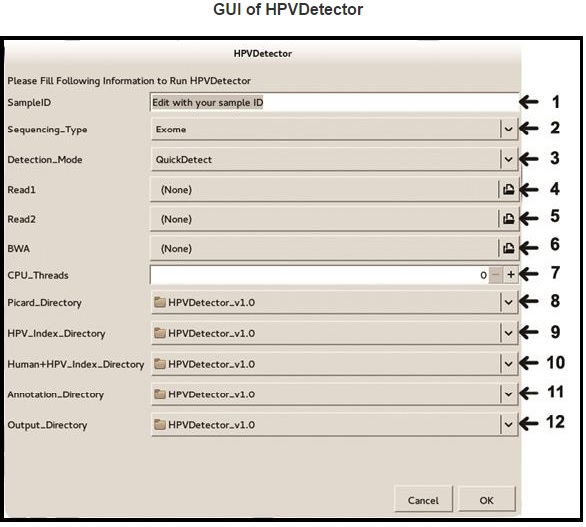
Upon successful execution, HPVDetector GUI will pop-up with 12 options for user selection. User can easily select required options using point-and-click interface and clicking on OK button will start running HPVDetector.
Step-1: Provide the sample ID
Step-2: Select sequencing type from Exome, Genome or Trenscriptome
Step-3: Select detection mode from QuickDetect or Integration
Step-4: Select input fastq file (forward reads fastq)
Step-5: Select input fastq file (reverse reads fastq)
Step-6: Select BWA executable
Step-7: Set the number of CPU threads as per your system (most modern CPUs have at least two cores/threads)
Step-8: Select directory where Picard Tools are installed
Step-9: Select directory where HPV_index is stored
Step-10: Select directory where Human-HPV_index is stored
Step-11: Select directory where Human-HPV Annotation files are stored
Step-12: Select a directory where output files should be stored
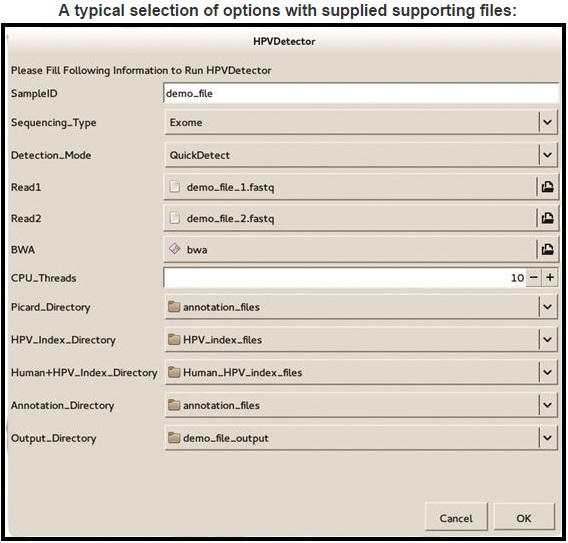
After selecting appropriate options, program can be executed by clinking on OK button. Progress can be monitored on screen and after completion, program will point to respective output files.