An integrated approach to determine the abundance, mutation rate and phylogeny of the SARS-CoV-2 genome.
Desai S, Rashmi S, Rane A, Dharavath B, Sawant A, Dutt A.
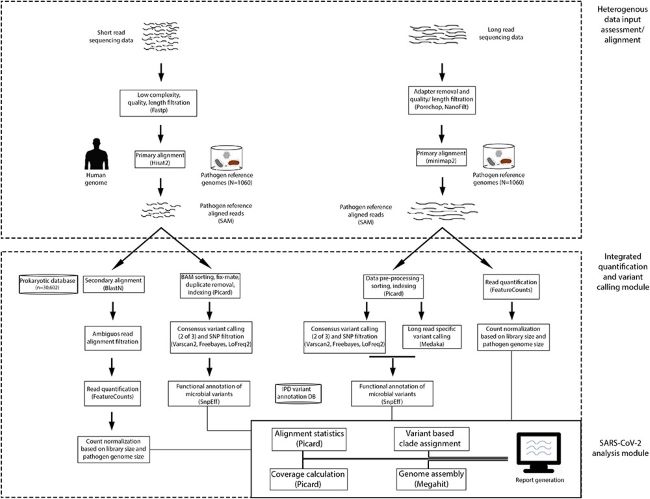
Next-generation sequencing (NGS) techniques have been widely used in the current Covid-19-pandemic to study various aspects of the SARS-CoV-2 genome dynamics. Global consortia efforts are aimed at extensive sequencing of the viral samples across populations, in search for therapeutic and preventive intervention avenues. However, plurality in the technology of sequencing generates diverse NGS data, requiring specialized analysis platforms. This presents a formidable computational challenge in public health laboratories engaged in studying epidemic outbreaks, thus limiting its application. To address this, we have developed Infectious Pathogen Detector (IPD), a graphical user interface (GUI) based automated quantification, variant and phylogenetic analysis pipeline for 1060 pathogens from heterogeneous NGS data.
We demonstrate application of IPD, analysing 1500 short- and long-read SARS-CoV-2 sequencing datasets, including pre-COVID-19 pandemic probands. The pathogen quantitation module could identify a varying burden (5.05 - 999655.7 FPM) of SARS-CoV-2 transcripts from the pandemic probands, while MERS and other pathogen reads were found in the pre-pandemic probands in the range of 22.7 - 455997.7 FPM. The IPD based variant analysis on the SARS-CoV-2 positive samples could identify 4634 SARS-CoV-2 variants (~ 3.05 per sample) across the genome with hotspot mutations in the ORF1ab and S gene, as reported earlier. Using the variants identified, we further performed phylogenetic clade assignment to demonstrate utility of IPD in performing isolate/ strain tracing. IPD is enabled with a GUI, allowing researchers to use it without any prior computational know-how, to generate an automated report for individual or bulk samples. IPD is freely available for download at: http://www.actrec.gov.in/pi-webpages/AmitDutt/IPD/IPD.html.
In summary, we present a GUI based, integrated pathogen analysis pipeline IPD and demonstrate its utility by analysing several publicly available SARS-CoV-2 genomic datasets. The IPD predicts the occurrence and dynamics of variability among infectious pathogens—with a potential for direct utility in the COVID-19 pandemic and beyond, to help automate the NGS based pathogen analysis and in responding to public health threats, efficaciously.