A one-step, one-tube real-time RT-PCR based assay with an automated analysis for detection of SARS-CoV-2. Heliyon.
Dharavath B, Yadav N, Desai S, Sunder R, Mishra R, Ketkar M, Bhanshe P, Gupta A, Redhu AK, Patkar N, Dutt S, Gupta S, Dutt A.
In this study, we describe a blueprint for academic centres, a rapid, easy to implement, validated real-time PCR based assay to detect SARS-CoV-2. The assay is complemented with automated analysis using an in-house developed novel platform-independent COVID qPCR Analyzer tool with graphical user interface (GUI). The tool analyses the raw qRT-PCR data in an unbiased manner, to enable in-house testing across laboratories to detect SARS-CoV-2.
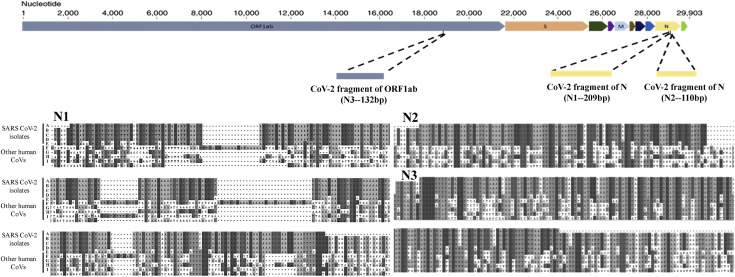
We first identified a unique conserved region in the Nucleocapsid gene (N1) and confirmed its in silico specificity by sequence comparison with human and other SARS-like viral genomes. We cloned and generated synthetic construct for the selected region, in addition to two literature reported SARS-CoV2 genomic regions; one each from Nucleocapsid (N2) and ORF1ab gene (N3). The three synthetic fragments (N1, N2 and N3) were spiked in the human DNA and RNA at varying concentrations for comparison of the detection limit of probe-free SYBR Green dye and multiplex TaqMan probe-based RT-PCR assay. The probe-based assay was found to be more sensitive with detection of up to 15 RNA copies/ reaction, as compared to 150 RNA copies/ reaction of the probe-free method. Since the probe-based approach could also detect the multiple reference target of SARS-CoV-2 and internal reference human control genes in single reaction, this was further developed into a one-step, one-tube, multiplex diagnostic kit, probing for the SARS-CoV2 Nucleocapsid and human RNase P internal control genes. Detection accuracy of the kit was evaluated with a panel of 26 clinical samples by performing 184 validations with dilutions of samples ranging from 1-100 ng. The kit was found to be 100% specific and 100% sensitive based on accurate identification of 5 of 26 positive samples and calling rest 21 negative samples as negative. For individual reaction, the kit costs under $3 and has a turnaround time of less than 2 hours, allowing quick and low-cost detection of SARS-CoV-2 from human samples.
Furthermore, to minimize variability and automate the qRT-PCR downstream analysis of multiple samples in the same run, we developed an open access freely available GUI based analytical tool, COVID qPCR Analyzer (www.actrec.gov.in/pi-webpages/AmitDutt/Covid/Analyzer.html), to detect the virus in an unambiguous and reproducible manner with minimal requirement of third-party tools on a platform-independent basis, for its universal application across all kinds of real-time PCR machines.
In summary, we present a rapid, easy to implement, real-time PCR based assay with automated analysis using a novel platform-independent COVID qPCR Analyzer tool with GUI to analyze the raw qRT-PCR data in an unbiased manner at a cost of under $3 per reaction and turnaround time of less than 2h, to enable in-house testing across laboratories to detect SARS-CoV-2. Given the immediate applicability of the method presented in this manuscript that may help to lower the transmission rate of the current COVID-19 pandemic, I would appreciate a fast review, if possible.