TMC-SNPdb 2.0: an ethnic-specific database of Indian germline variants.
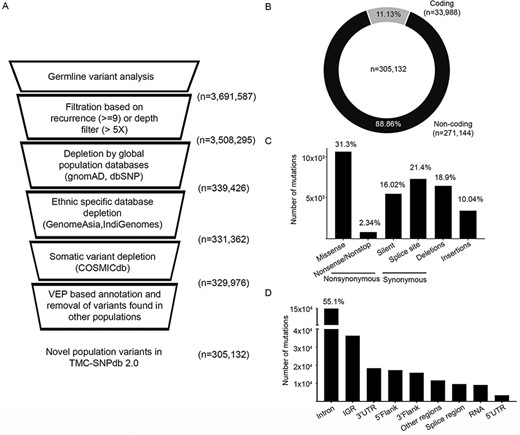
Desai S, Mishra R, Ahmad S, Hait S, Joshi A, Dutt A.We earlier described (TMC-SNPdb – DOI:10.1093/database/baw104, Upadhyay et. al. 2016) with 1,14,309 Indian-specific variants that have been downloaded by 131 labs across the globe, to date. Here, in this manuscript, we present an updated version of the Indian germline variant database, TMC-SNPdb 2.0, with a GUI-enabled, biologist friendly, accompanying toolkit to integrate the resource in the somatic analysis pipeline, as the most exhaustive open-source reference database of germline variants predominantly occurring across 1800 Indian individuals. The GUI-based toolkit also allows researchers to create their own normal variant database, integrating various germline database resources and annotation of variants for their presence in the germline variant database.
Specifically, we integrate the recent sequencing efforts undertaken by the Genomics for Public Health in India (IndiGen) program for 1029 healthy individuals, the GenomeAsia 100K initiative for 598 healthy individuals, along with our in-house efforts of 173 normal samples derived from cancer patients of Indian origin to present TMC SNPdb 2.0. From the analysis of 173 normal exome samples derived from cancer patients, we identified 305,132 unique variants. The majority of the variants were obtained from the non-coding region of the genome (88.86%, n=271144), whereas 11.13% (n=33988) were within the coding region. Among the coding region, we identified 10614 missense variants which would be specifically labeled as “novel” or “variants of unknown significance” in any of the somatic analyses.
Integrating the publicly available Indian population variant data from IndiGenomes and GenomeAsia, with TMC-SNPdb 2.0, we further demonstrate its utility by analyzing the whole-exome sequence from 224 in-house tumor samples (180 paired and 44 orphans) of Indian origin. We show an additional average depletion of 3.44% variants per paired tumor and significantly higher (p-value<0.001) for orphan tumors (4.21%), demonstrating the utility of the rare, unique variants found in the ethnic-specific variant datasets in reducing the false positive somatic mutations.
Overall, TMC SNPdb 2.0 is the most exhaustive open-source reference database of germline variants occurring across 1800 Indian individuals to analyze cancer genomes and other genetic disorders.
The database and toolkit package is available for download at http://www.actrec.gov.in/pi-webpages/AmitDutt/TMCSNPdb2/TMCSNPdb2.html.