
HPVDetector-an open source tool for biologists
Human papilloma virus (HPV) accounts for the most common cause of all virus-associated human cancers. However, despite large-scale genome wide DNA sequencing efforts of the cancer genome there is no dedicated informatics tool to rapidly detect the presence of HPV in these genomes, in an exclusive manner. There are indeed a variety of gene integration finding tools available that can detect different pathogen insertions in the human genome such as ViralFusionSeq, VirusSeq, VirusFinder, Path-Seq, RINS, and ReadSCAN. These sophisticated tools though have their specific third party needs, necessitate intense computational infrastructure, cannot be run without specialized and advanced computational expertise of the researcher, and more impotantly are not specific for HPV detection, per se—for e.g., lacks information to annotate the region of HPV genome to predict the integrated viral gene, of which some are know to function as oncogenes.
In this study, we present a novel freely distributable computational tool “HPVDetector” (through a weblink download) to detect all known HPV types along with their sites of integration in the host genome using next generation sequencing data set, along with a widely compatible annotated reference of 143 HPV genome as a resource. This user friendly tool has been designed for researchers who has limited computational expertise using graphical user interface (GUI) that requires minimal third party tools. Using HPVDetector, one can analyze paired end whole exome, whole genome or whole transcriptome dataset to detect all known HPV types along with their sites of integration in the host genome. The tool can run in two modes: a quick detect mode can identify co-infection of HPVs and their quantitative abundance while integration mode can identify HPV integration loci in human genome and provide comprehensive HPV specific annotations. Based on our evaluation with 78 exome, 23 transcriptome and 1 whole genomes, HPVDetector was able to identify presence of HPV in 17 exome and 4 transcriptome data. Using the annotation module, we could show that viral gene E7 was most widely rerpresented among all the reads detected that is a known viral oncogene. Additionally, the integration module allowed us to validate known HPV integration sites, identify known fragile sites of the human genome as HPV integration site and novel integration sites. We also present a lucid step wise instructions as a supplementary info along with other details directed towards non computational biologsts to use the tool.
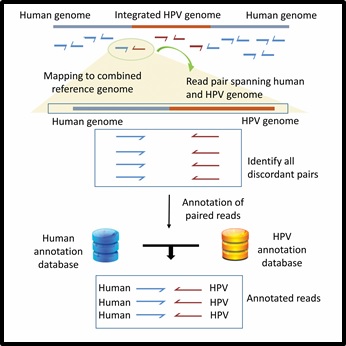
[Conceptual workflow of the HPVDetector
The flowchart represents workflow for HPVDetector. Paired end reads obtained from Next generation sequencing data are aligned to a combined Human-HPV reference database. All discordant read pairs with one read aligning to human and other to the HPV genome are identified and annotated utilizing human and HPV database using an inbuild annotator module.
]
[wellcomedbt]
[HPVDetector]