
First Indian germline SNPdb
Here’s a pilot attempt to fill the gap of lack of Indian specific dbSNP. We present “TMC-SNPdb”, as the first open source freely available, flexible and upgradable SNP database from whole exome data of 62 normal tissues derived from cancer patients (hosted at ANNOVAR and dbSNP149). Following stringent computational subtraction method, TMC-SNPdb consists of 114, 309 unique germline variants prevalent exclusively among Indian population at variant frequency. TMC-SNPdb comes along with a companion subtraction tool that can be executed with command line option or using an easy-to-use graphical user interface (GUI) to deplete Indian population specific SNPs, in addition to the dbSNP and 1000 Genomes Project.
We show the utility and effectiveness of the tool by analyzing whole exome data set of 132 cancer samples of Indian origin. We demonstrate that “TMC-SNPdb” significantly reduced 42%, 33% and 28% false positive somatic events, post dbSNP depletion, in head and neck cancer, gallbladder cancer and cervical cancer samples, respectively. This is the first report of such exhaustive SNPs from Indian population based on next generation sequencing data as a crucial resource to the community not restricted only to the Indian population, that will find its application for researchers studying various human diseases.
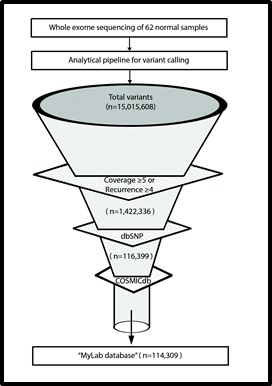
[Development of TMC-SNPdb using whole exome sequencing: Schematic flow representation of steps followed. Whole exome sequence of 62 normal tissue were analysed using GATK to generate vcf files. Raw variants obtained were further filtered to find list of variants absent in dbSNP and COSMICdb.]